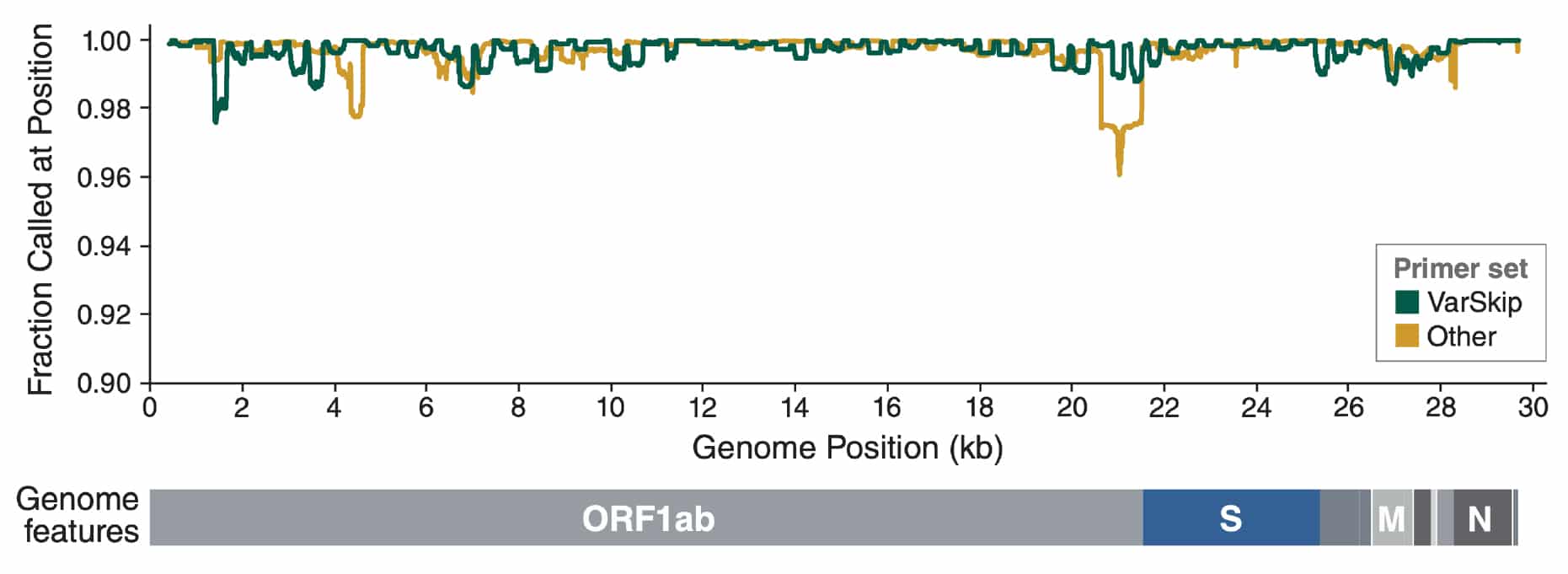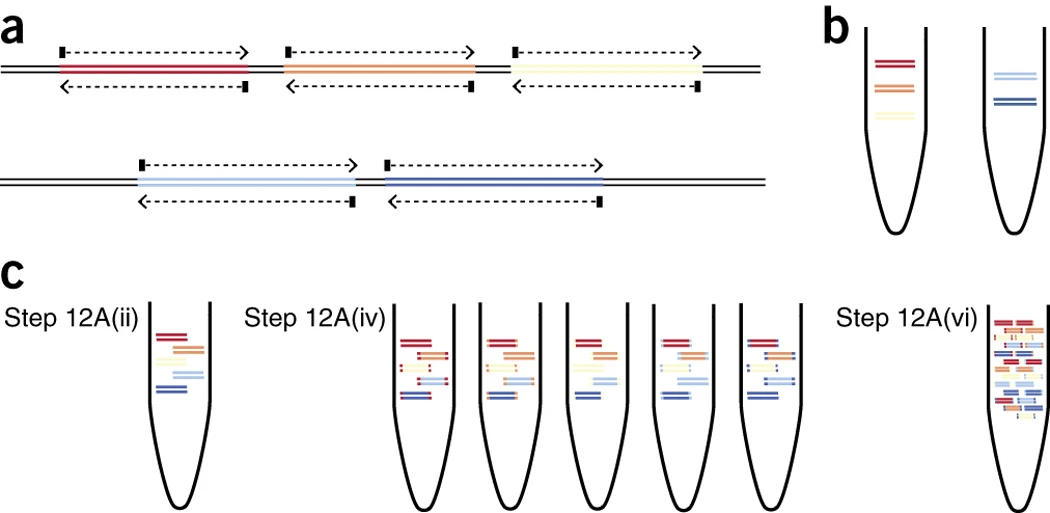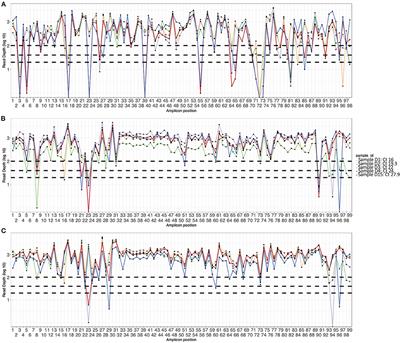
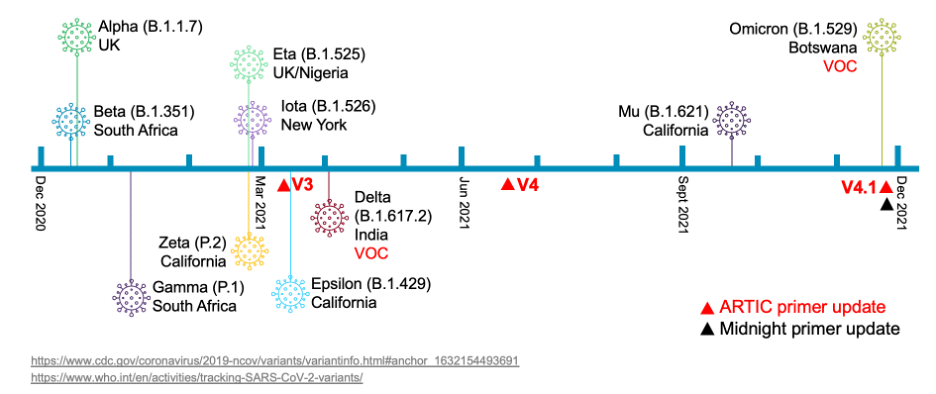
Frontiers | Optimization of the SARS-CoV-2 ARTIC Network V4 Primers and Whole Genome Sequencing Protocol
Omicron and beyond: HiFiViral Kit provides labs with a future-proof solution for emerging COVID-19 variants - PacBio
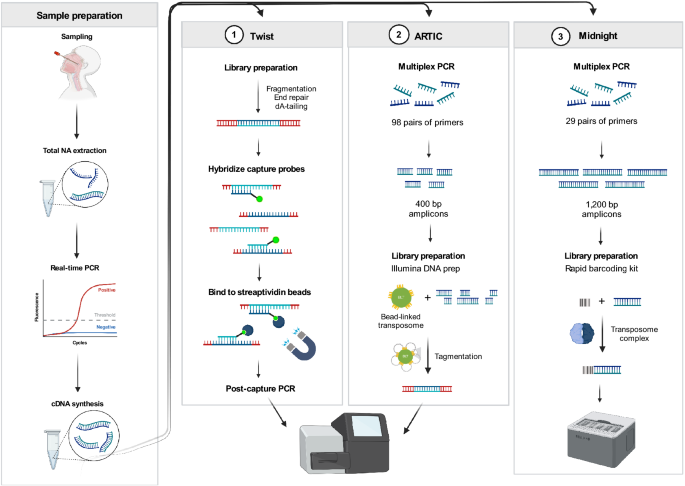
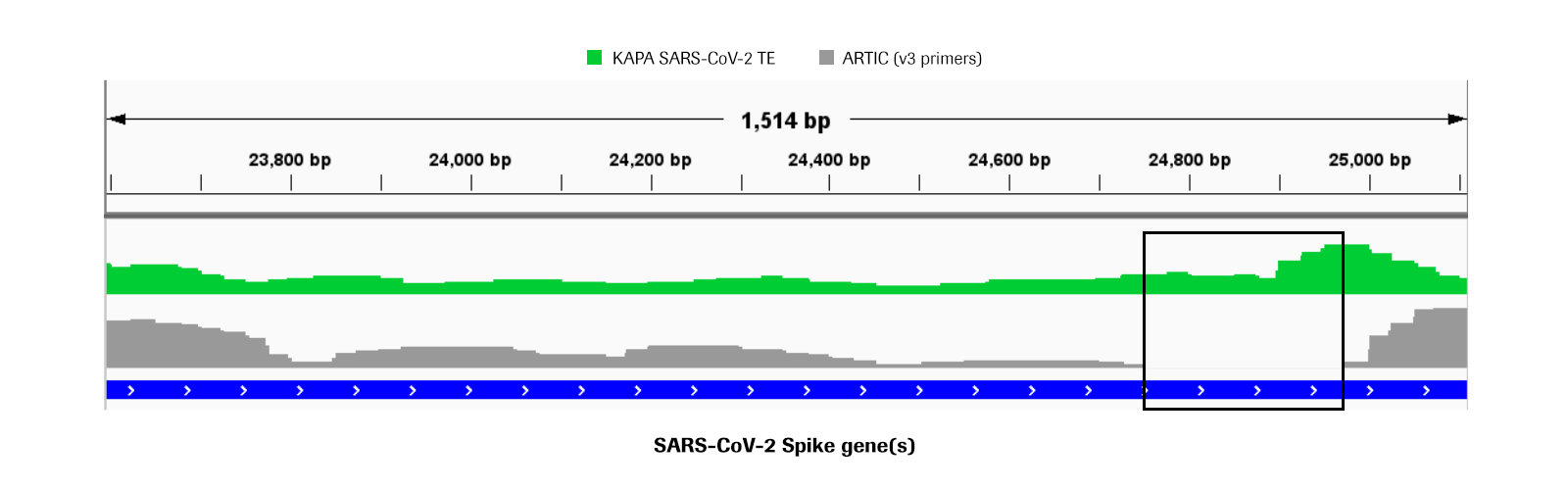
Comparison of SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization | Scientific Reports
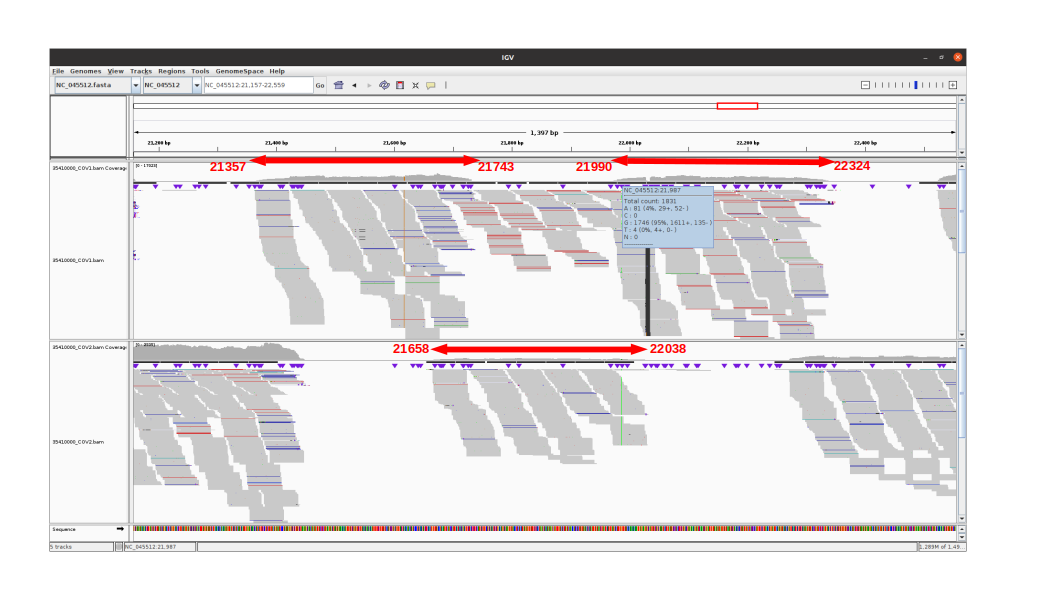
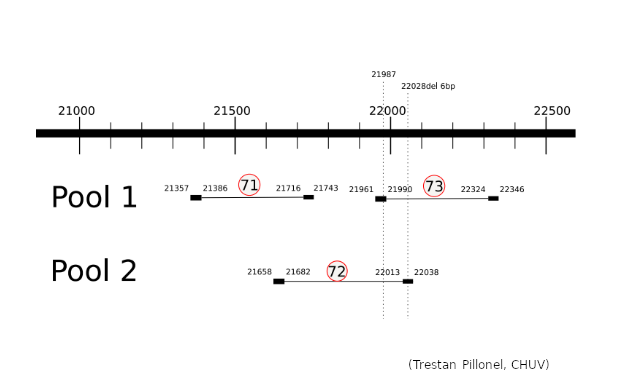
Missing G21987A mutation in SARS-CoV-2 delta variants due to non-specific amplification by ARTIC v3 primers - nCoV-2019 Genomic Epidemiology - Virological

New England Biolabs on Twitter: "We can all agree on the need for reliable, accurate & fast methods for sequencing SARS-CoV-2. The #NEBNext ARTIC kits are based on the work of @NetworkArtic

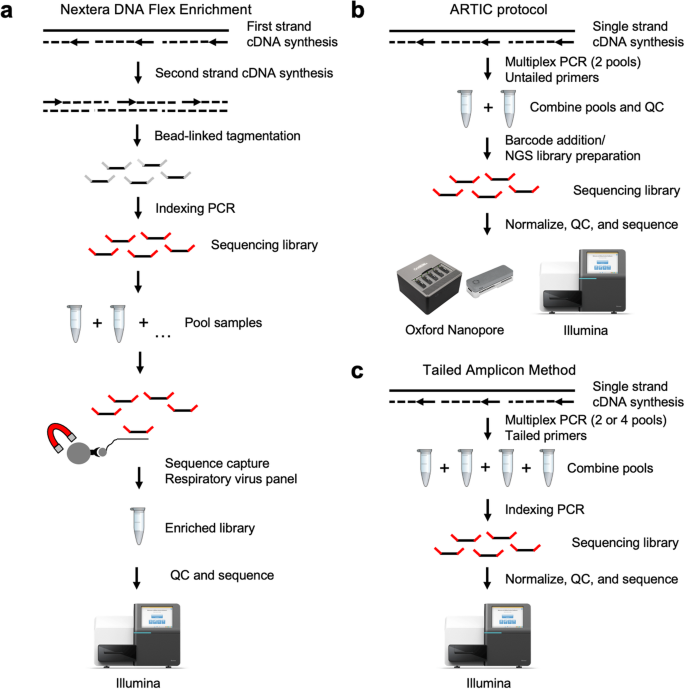
SARS-CoV-2 Genome Sequencing Methods Differ in Their Abilities To Detect Variants from Low-Viral-Load Samples | Journal of Clinical Microbiology
A short plus long-amplicon based sequencing approach improves genomic coverage and variant detection in the SARS-CoV-2 genome
Bioinformatics workflows for SARS-CoV-2; from raw Nanopore reads to consensus genomes using the ARTIC coronavirus protocol
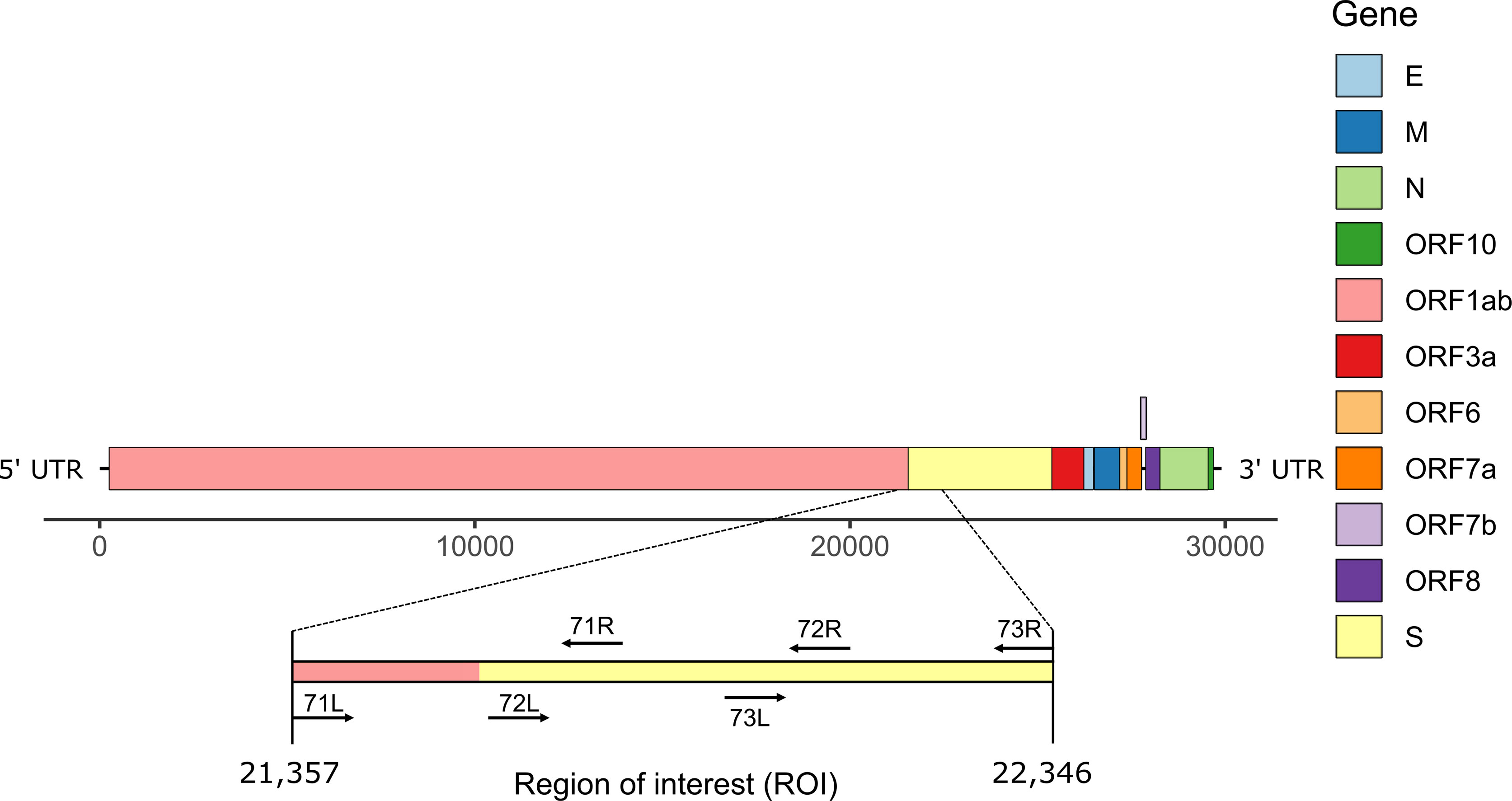
Frontiers | Investigating the Extent of Primer Dropout in SARS-CoV-2 Genome Sequences During the Early Circulation of Delta Variants

periscope: sub-genomic RNA identification in SARS-CoV-2 ARTIC Network Nanopore Sequencing Data | bioRxiv

ARTIC Network provides protocol for rapid, accurate sequencing of novel coronavirus (nCoV-2019): first genomes released

Variation at Spike position 142 in SARS-CoV-2 Delta genomes is a technical artifact caused by dropout of a sequencing amplicon | medRxiv
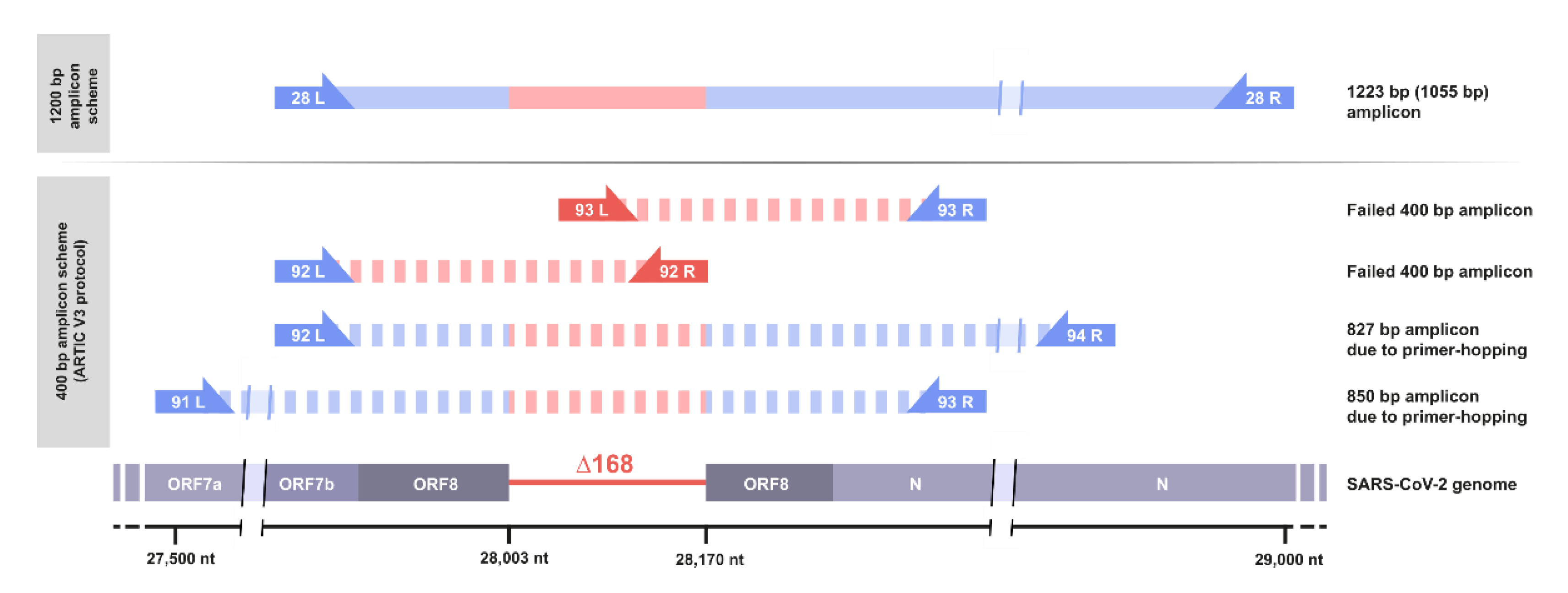
Viruses | Free Full-Text | Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling Pipelines
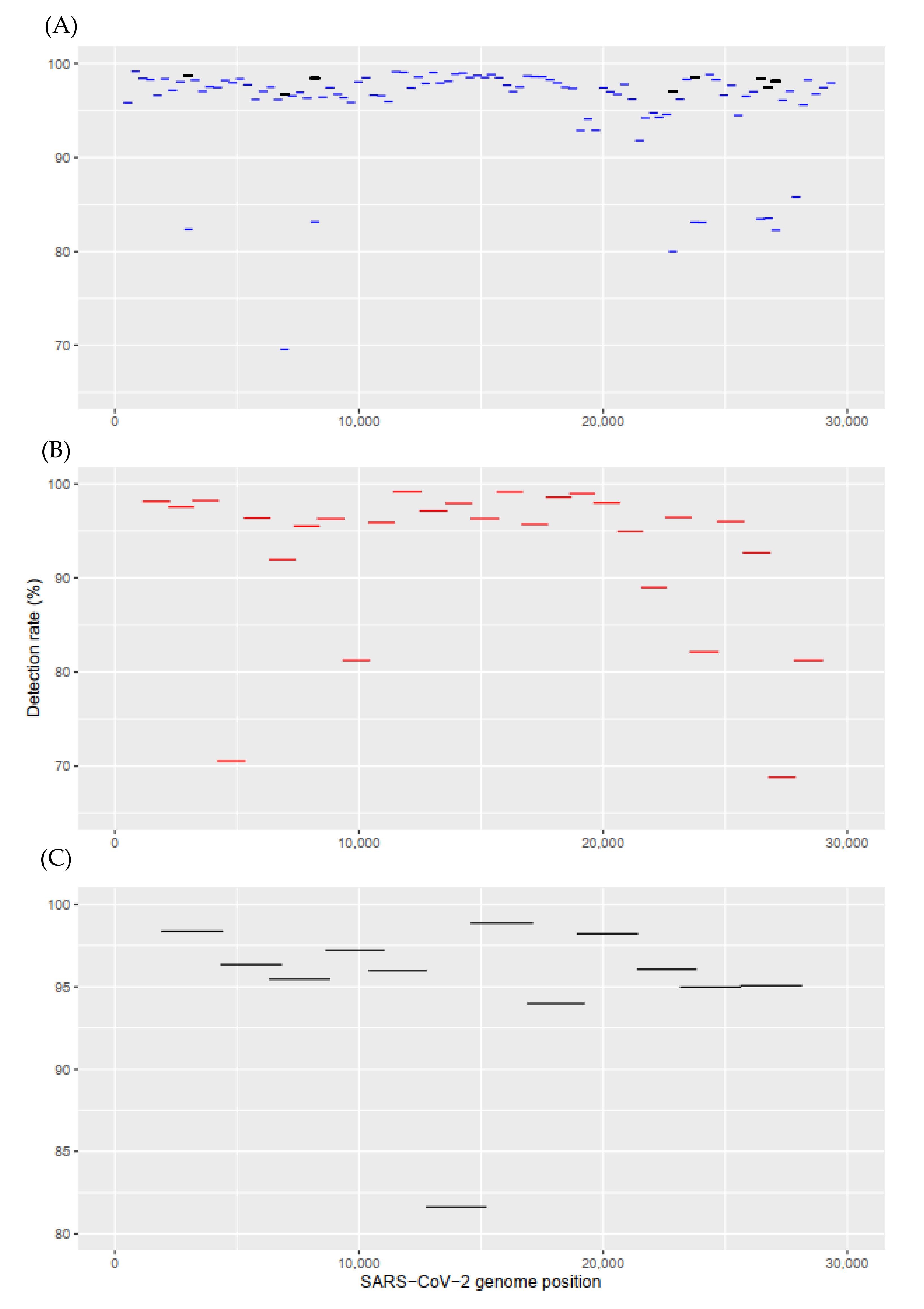
IJMS | Free Full-Text | High-Integrity Sequencing of Spike Gene for SARS-CoV-2 Variant Determination
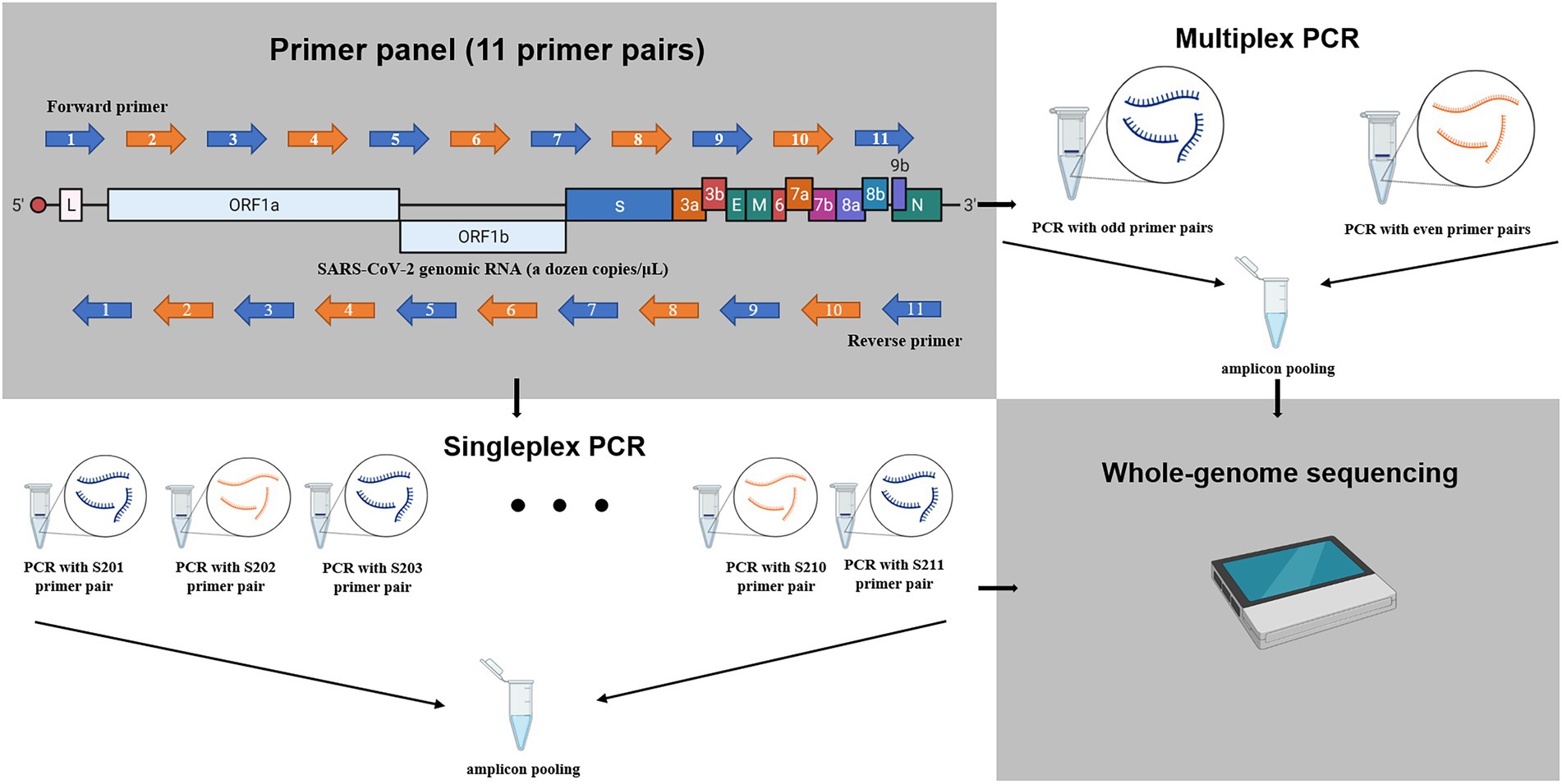
A proposal of alternative primers for the ARTIC Network's multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing